IDRMutPred - Help
1. Introduction
IDRMutPred is developed for identifying nsSNVs as disease-associated or neutral in IDRs. Based on 17 optimal features and the training set containing only IDR nsSNVs, we trained IDRMutPred by using LightGBM, XGBoost, and Random Forest, and it outperforms other general predictors when evaluating on testing datasets containing only IDR nsSNVs. IDRMutPred will be valuable in the studies related to intrinsically disordered proteins and/or precision medicine.
2. Predictor Input
IDRMutPred predictor is as below:
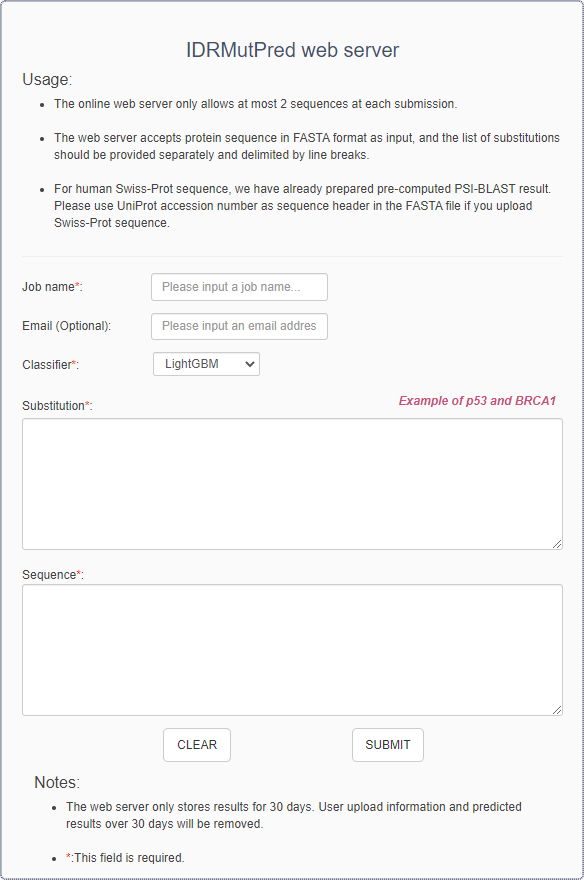
- The job name could be composed by character, digit, underline, vertical bar, dot, and square brackets and its length < 20.
- Leave an email address for receiving the prediction results. The server will check if you have entered a valid address.
- As default, IDRMutPred predictor will use LightGBM as default classifier, but user can choose one of three classifier (LightGBM, XGBoost, RandomForest) as backend classifier.
- Input the list of amino acid substitutions. Substitutions should be provided separately and delimited by line breaks.
- Input the protein sequences in standard FASTA format. The unnatural amino acids are not allowed. The maximum input is 2 sequences.
- Click the button “SUBMIT” to submit the job.
The IDRMutPred predictor will take some time to run the job. Before the job complete, the result page will show as following:

3. Predictor Output
When the job is completed, the result page will present as following:
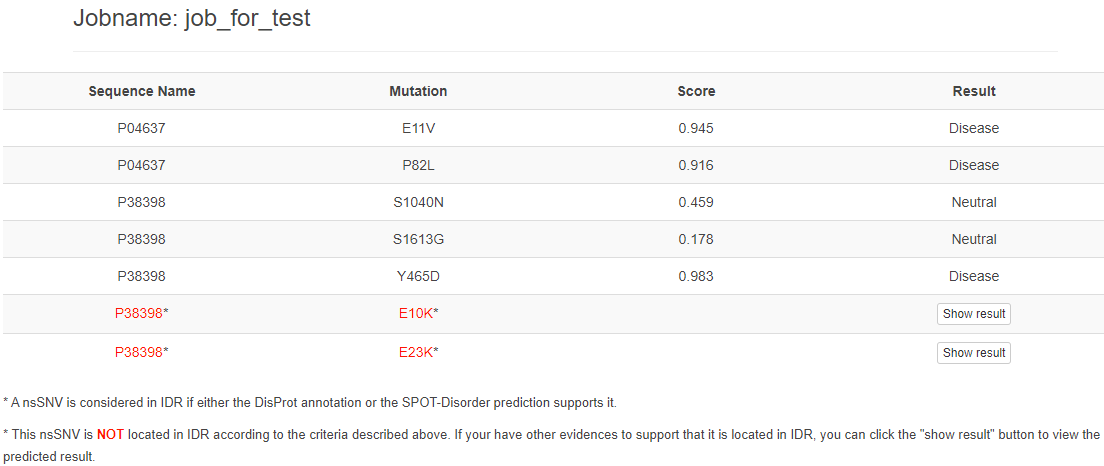
The criteria for determine whether a nsSNV is located in IDR are provided by SPOT-Disorder and DisProt annotations.If the nsSNV is not located in IDR, it would be marked in red.
If your're sure this nsSNV located in IDR, please click button to show the result:
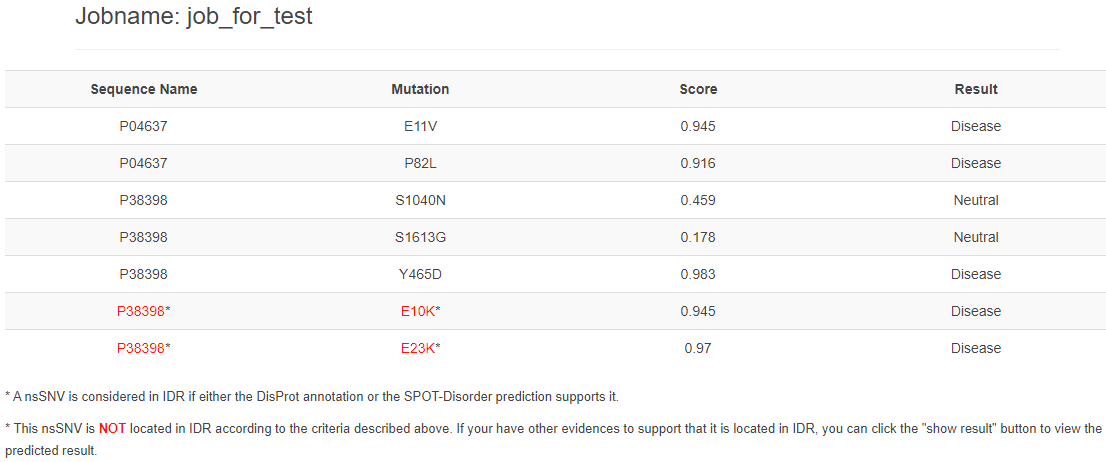
4. Citation
5. What's new
September 15, 2019: Initial version of web server is released now! Welcome feedback.
September 25, 2019: Now users can download standalone package of IDRMutPred.
June 21, 2020: Now users can download docker image of standalone package.