Introduction
WDSPdb 3.0
- WDSPdb, originally created in 2014, is a database for WD40-repeat proteins. It serves as a free and comprehensive resource to provide high quality structural information of WD40 proteins as well as the featured sites, including the special hydrogen-bond network residues and interaction hotspot residues. WDSPdb 3.0 is based on UniprotKB 2022_02, and it stores more than 1,600,000 predicted WD40-repeat proteins from about 36,000 species. Compared to its previous version (WDSPdb 2.0, released in 2019), the proteins in WDSPdb 3.0 have increased about 1,000,000.
- WDSPdb provides accurate structure predictions and featured sites annotations specifically for WD40-repeat domains, based on our in-house prediction tool, WDSP. During the updating of this release, WDSP has been configured to use Psipred 4.0.1, and NCBI BLAST+ 2.13.0 against Uniref90(Release_202202), and 3 iterations with random number seeds of 0,1,2.
- It exhibited the 3D structure models for WD40-repeat proteins whose repeat counts are 6, 7 or 8, and whose confidence category is 'High'. These 3D structures were modeled by MODELLER 10.4 using our customized target-template alignments.
- It mapped about 28,000 missense variants/mutations/substitutions to 231 human WD40 proteins from Swiss-Prot section with confidence category of 'High'. These data are from 9 sources: CancerHotspots, COSMIC, Clinvar, Humsavar from UniprotKB, 1000 Genomes phase 3, IntoGen, IntAct, ExAC, and GnomAD exomes. Besides the general information of variants, we not only provided the special annotations include exact secondary structure locations, featured site identifications (hydrogen bond network, potential hotspots on the top face), but also presented the information obtained from other databases, like pathogenic predictions in dbNSFP,cancer driver mutation from the IntOgen, cancer highly recurrent mutations from the CancerHotspots, and whether the variants were experimentally shown to affect the PPIs based on IntAct annotations.
- The updated WDSP prediction tool that was adopted for building WDSPdb 3.0 is also deployed as a web server. If the interested sequence is not included in the database, the user can submit it to this online tool to make predictions.
WD40 Repeat Protein
- The WD40-repeat domains are one of the most abundant interactors in protein-protein interaction (PPI) networks. By acting as scaffolds, they assemble various molecular machineries, and play versatile roles in fundamental biological processes including signal transduction, expression regulation, histone modification, ubiquitination, cell cycle control, etc.
- In structure, WD40 domain is a β-propeller usually formed by 6-8 blades, and each blade contains 40-60 residues with conserved GH and WD dipeptides.
- Each WD40 blade is a four-stranded antiparallel β-sheet (strand a, b, c and d, connected by loops), and is often stabilized by a strong side-chain hydrogen bond network that is widely and uniquely presented in WD40 proteins. These hydrogen-bond network residues servers as a group of featured sites of WD40 domains.
- The structural blades correspond to the repeated units in sequence, with one strand shifted. That is, a blade contains strand a, b, c, d consecutively, while a repeat unit contains strand d of the previous blade, and strand a, b, c of the blade that follows.
- WD40 domain has three faces, i.e. top, side and bottom faces, to mediate the interactions with other molecules. The top face is better studied than others, and the potential hotspot residues on this face are exposed into the solvent by the β bulge between the strand a and strand b to participate in interactions.
WDSP
- WD40 repeat protein Structure Predictor (WDSP) was developed to accurately predict the secondary structures of WD40 domains, featured sites including hydrogen bond network residues, and interaction hotspot residues.
- The WDSP tool adopts a WD40-specific position weight matrix (PWM) and PSIPRED as backends.
- WDSP is the only tool which can both identify the exact boundaries of WD40 repeat and their structural and functional features up to date.
Statistics
The WDSPdb 3.0 is based on UniProtKB 202202, and the data coverage has increased to about 1.6 times of WDSPdb 2.0.
The statistic of all WD40 proteins in different categories
| Category | WD40 Proteins | WD40 Repeats | Hydrogen Bond Network | Hotspots on the Top Face |
|---|---|---|---|---|
| Total | 1,670,938 | 12,617,145 | 2,611,632 | 15,660,903 |
| High | 576,687 | 4,776,558 | 2,154,765 | 6,427,038 |
| Middle | 203,996 | 1,377,451 | 297,679 | 1,649,854 |
| Low | 890,255 | 6,463,136 | 159,188 | 7,584,011 |
| Eukaryota | 1,067,648 | 7,887,352 | 2,275,636 | 9,961,239 |
| Bacteria | 572,434 | 4,499,652 | 325,898 | 5,432,329 |
| Archaea | 21,672 | 173,014 | 7,645 | 198,585 |
| Viruses | 942 | 6,550 | 332 | 8,622 |
| Homo sapiens (9606) | 2,120 | 13,569 | 3,932 | 17,099 |
| Mus musculus (10090) | 1,362 | 9,620 | 2,758 | 12,118 |
| Rat (10116) | 1,062 | 8,500 | 2,616 | 10,745 |
| Frog (8355) | 1,682 | 13,446 | 4,008 | 17,241 |
| Danio rerio (7955) | 1,122 | 8,264 | 2,304 | 10,389 |
| Drosophila melanogaster (7227) | 621 | 4,634 | 1,305 | 5,944 |
| C. elegans (6239) | 311 | 2,296 | 614 | 2,889 |
| Oryza sativa (39947) | 947 | 6,425 | 2,077 | 8,097 |
| Arabidopsis thaliana (3702) | 1,649 | 12,600 | 3,864 | 15,786 |
| Saccharomyces cerevisiae (559292) | 142 | 1,154 | 310 | 1,513 |
| Schizosaccharomyces pombe (284812) | 160 | 1,294 | 380 | 1,657 |
Swiss-Prot
The statistic of Swiss-Prot section WD40 proteins in different categories
| Category | WD40 Proteins | WD40 Repeats | Hydrogen Bond Network | Hotspots on the Top Face |
|---|---|---|---|---|
| Total | 1,667,054 | 12,587,025 | 2,602,545 | 15,621,282 |
| High | 574,455 | 4,759,298 | 2,146,262 | 6,402,584 |
| Middle | 203,715 | 1,375,068 | 297,304 | 1,647,156 |
| Low | 888,884 | 6,452,659 | 158,979 | 7,571,542 |
| Eukaryota | 1,064,331 | 7,860,906 | 2,266,699 | 9,926,306 |
| Bacteria | 571,902 | 4,496,222 | 325,755 | 5,427,986 |
| Archaea | 21,663 | 172,949 | 7,644 | 198,493 |
| Viruses | 916 | 6,371 | 326 | 8,369 |
| Homo sapiens (9606) | 1,726 | 10,266 | 2,937 | 12,961 |
| Mus musculus (10090) | 1,000 | 6,493 | 1,854 | 8,157 |
| Rat (10116) | 930 | 7,393 | 2,239 | 9,307 |
| Frog (8355) | 1,587 | 12,684 | 3,747 | 16,233 |
| Danio rerio (7955) | 1,029 | 7,596 | 2,062 | 9,508 |
| Drosophila melanogaster (7227) | 528 | 3,848 | 1,116 | 4,906 |
| C. elegans (6239) | 218 | 1,566 | 410 | 1,936 |
| Oryza sativa (39947) | 915 | 6,135 | 1,970 | 7,683 |
| Arabidopsis thaliana (3702) | 1,461 | 11,134 | 3,353 | 13,892 |
| Saccharomyces cerevisiae (559292) | 0 | 0 | 0 | 0 |
| Schizosaccharomyces pombe (284812) | 0 | 0 | 0 | 0 |
TrEMBL
The statistic of TrEMBL section WD40 proteins in different categories
| Category | WD40 Proteins | WD40 Repeats | Hydrogen Bond Network | Hotspots on the Top Face |
|---|---|---|---|---|
| Total | 1,667,054 | 12,587,025 | 2,602,545 | 15,621,282 |
| High | 574,455 | 4,759,298 | 2,146,262 | 6,402,584 |
| Middle | 203,715 | 1,375,068 | 297,304 | 1,647,156 |
| Low | 888,884 | 6,452,659 | 158,979 | 7,571,542 |
| Eukaryota | 1,064,331 | 7,860,906 | 2,266,699 | 9,926,306 |
| Bacteria | 571,902 | 4,496,222 | 325,755 | 5,427,986 |
| Archaea | 21,663 | 172,949 | 7,644 | 198,493 |
| Viruses | 916 | 6,371 | 326 | 8,369 |
| Homo sapiens (9606) | 1,726 | 10,266 | 2,937 | 12,961 |
| Mus musculus (10090) | 1,000 | 6,493 | 1,854 | 8,157 |
| Rat (10116) | 930 | 7,393 | 2,239 | 9,307 |
| Frog (8355) | 1,587 | 12,684 | 3,747 | 16,233 |
| Danio rerio (7955) | 1,029 | 7,596 | 2,062 | 9,508 |
| Drosophila melanogaster (7227) | 528 | 3,848 | 1,116 | 4,906 |
| C. elegans (6239) | 218 | 1,566 | 410 | 1,936 |
| Oryza sativa (39947) | 915 | 6,135 | 1,970 | 7,683 |
| Arabidopsis thaliana (3702) | 1,461 | 11,134 | 3,353 | 13,892 |
| Saccharomyces cerevisiae (559292) | 0 | 0 | 0 | 0 |
| Schizosaccharomyces pombe (284812) | 0 | 0 | 0 | 0 |
Tutorial
Home page overview
You can view the slides online below if you can access google docs. Otherwise you need to download the slides to view locally.
How to download data?
You can view the slides online below if you can access google docs. Otherwise you need to download the slides to view locally.
How to use search box?
You can view the slides online below if you can access google docs. Otherwise you need to download the slides to view locally.
Protein detail page
You can view the slides online below if you can access google docs. Otherwise you need to download the slides to view locally.
Manual
Home Page
- You can hit the “Home” button at the banner to quickly jump to the home page.
- You can hit the “Predictor” button at the banner to jump to the WDSP predictor page to preform the WD40 repeat prediction for your own sequences.
- You can use the “Help” page to learn how to use the database or predictor and understand the provided information. If you have any questions, please feel free to contact with us at the “Contact” page.
- You can click the "Categories" button on the banner to access the WD40 proteins belonging to different UniProt sections, different confidence categories, taxonomy classes, or different model organisms.
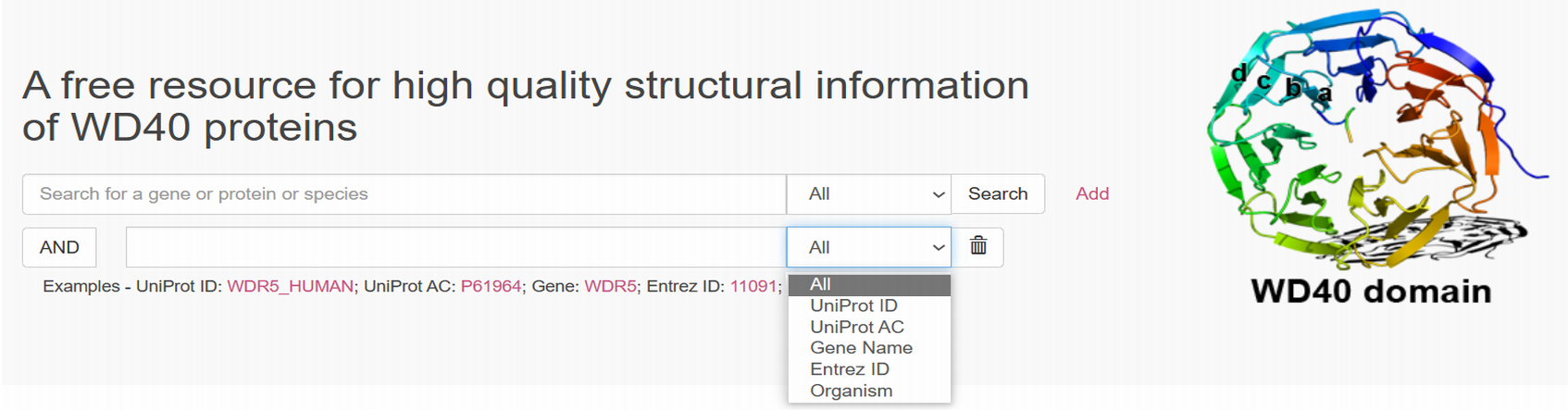
- You can get started by using the search bar at the top of the home page to search the proteins using search terms in different groups. There is a drop-down list that allows selecting the group, e.g., Uniprot ID, Uniprot AC, Gene name, Entrez ID, Organism. You can then input your search term(s) accordingly.
- You can also add multiple search bars via the "Add" button on the right. Then you can do advanced search by using multiple conditions.
- You can quickly learn about the WDSPdb and the introductions of WD40 repeat proteins, WDSP predictor, structural feature (hydrogen bond network), functional feature (hotspots on the top face).
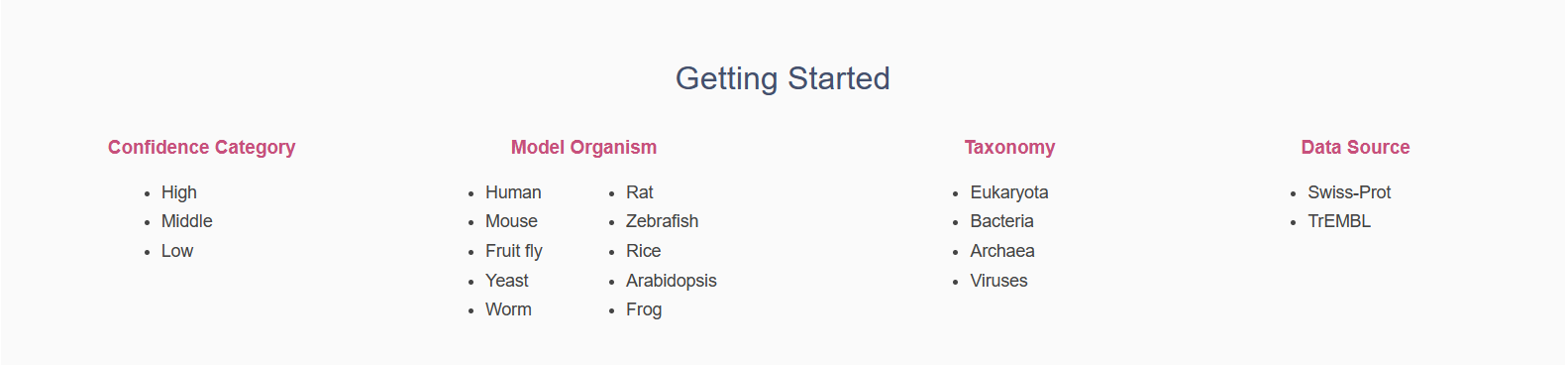
- Moreover, you can get started by browsing different classifications or organisms listed below the brief introductions.

- If you use the data of WDSPdb or the WDSP predictor in you research, please cite the associated papers.
Search Result Page
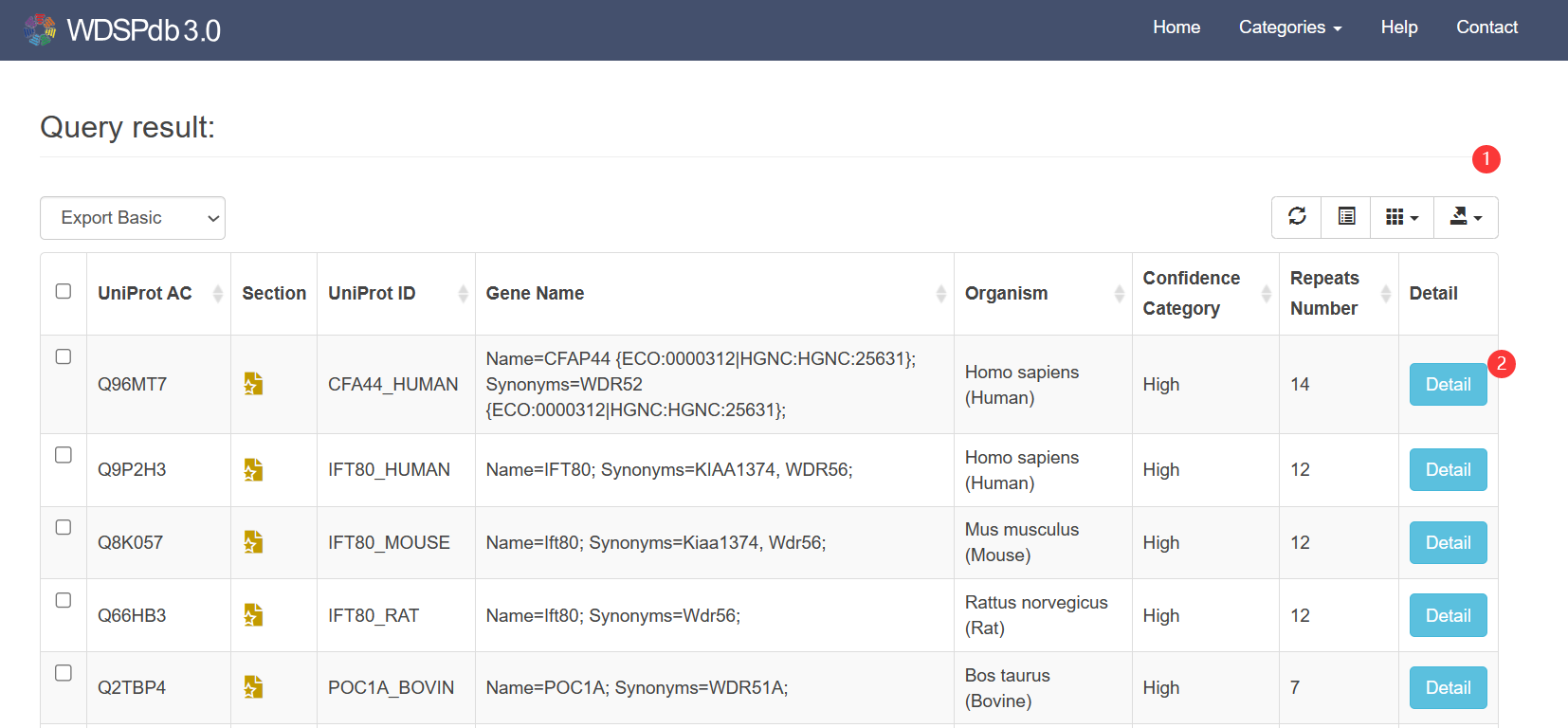
- The search results will be listed on a new page and sorted by "UniProt_AC".
- You can select specific columns and sort them according to the information of searched proteins.
- You can export the search results list or customized selected list by multiple formats, such as .txt, .csv, .json, .xml et al ①.
- You can click the "Detail ②" button to access the detailed page of your interested WD40 protein.
| Display by default | Columns | Explanation |
|---|---|---|
| On | Accession Number | UniProt accession number (can be sorted) |
| On | Section | UniProt section, Swiss-Prot or TrEMBL (cannot be sorted) |
| On | Entry Name | The UniProt entry name (can be sorted) |
| On | Gene Name | The name of the gene that encode this protein (can be sorted) |
| On | Organism | The Latin and English name of organism (can be sorted) |
| On | Confidence Category | The assigned confidence category for that protein as a WD40 protein (can be sorted) |
| On | Repeat Number | The predicted WD40 repeats number (can be sorted) |
| Off | WDSP Score | The total score of WDSP, i.e., sum of repeat scores, tetrad sores, and other scoring terms (can be sorted) |
Protein Detail Page
Basic protein information
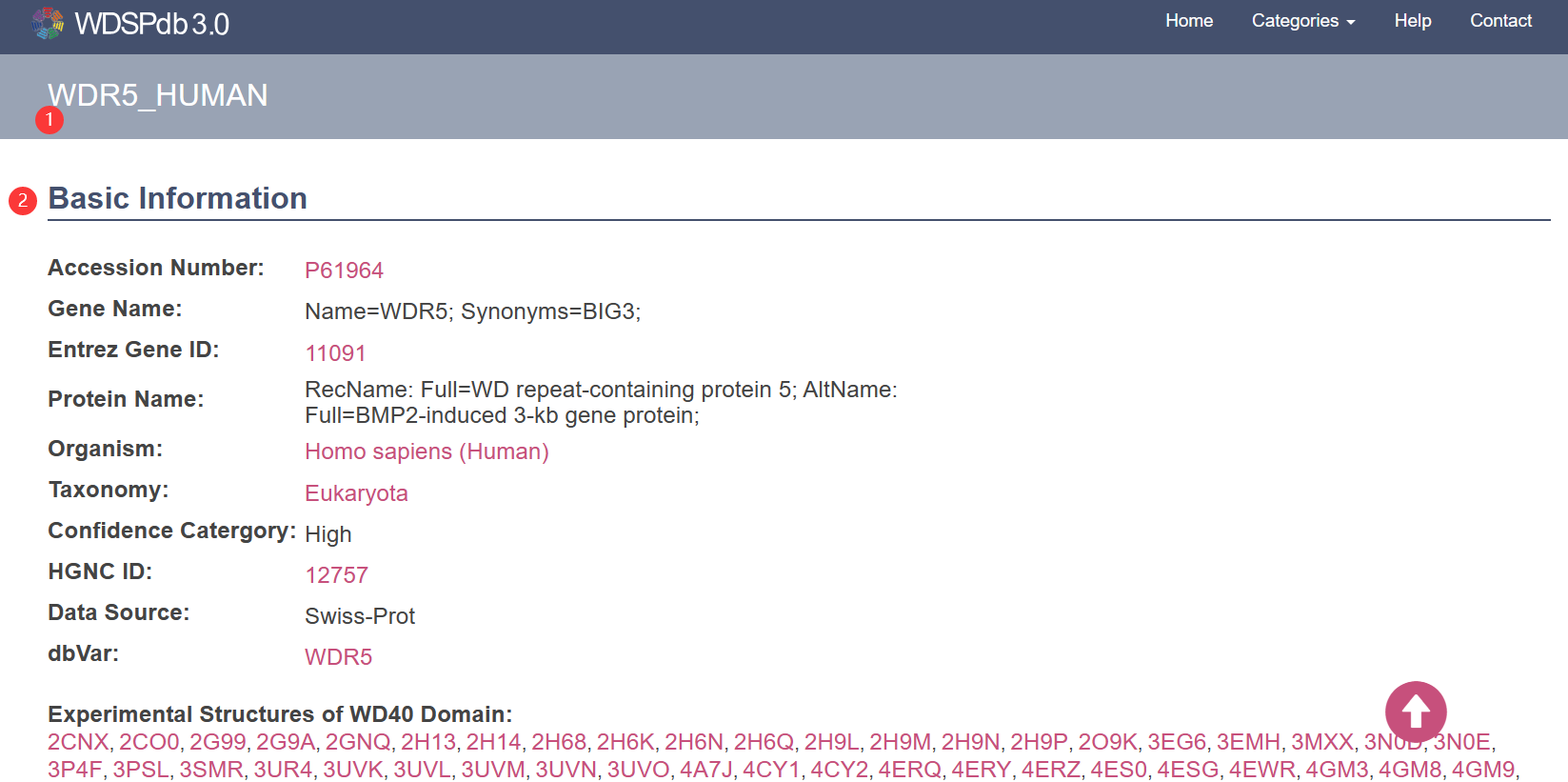
- Take WDR5_HUMAN ① information page as an example.
- The basic information of the protein is displayed as a table at the top of the page ②.
- The information items about include:
| Items | Explanation |
|---|---|
| Accession Number | UniProt accession number |
| Gene Name | The gene symbol |
| Gene ID | Entrez gene ID |
| Protein Name | Full protein name |
| Organism | The Latin and English name of organism |
| Organism Domain | The highest taxonomic group of organisms in the biological taxonomy. e.g., Eukaryota, Prokaryota, etc. |
| Confidence Category | The assigned confidence category for that protein as a WD40 protein |
| HGNC ID | The HGNC ID of the protein |
| Data Source | The protein belongs to Swiss-Prot or TrEMBL |
| dbVar | The link to dbVar for the gene encoding this protein if exists |
| Experimental Structures of WD40 Domain | The experimental structures covering one of the WD40 repeat |
| Reference Sequence | protein ID or transcript ID in NCBI Refseq database |
| Description | Alternative protein names |
| Functions | The annotated function from UniProt Knowledge Base |
| Ensembl ID | The Ensembl gene ID, transcript ID and protein ID of this protein |
The WD40 repeat and secondary structures
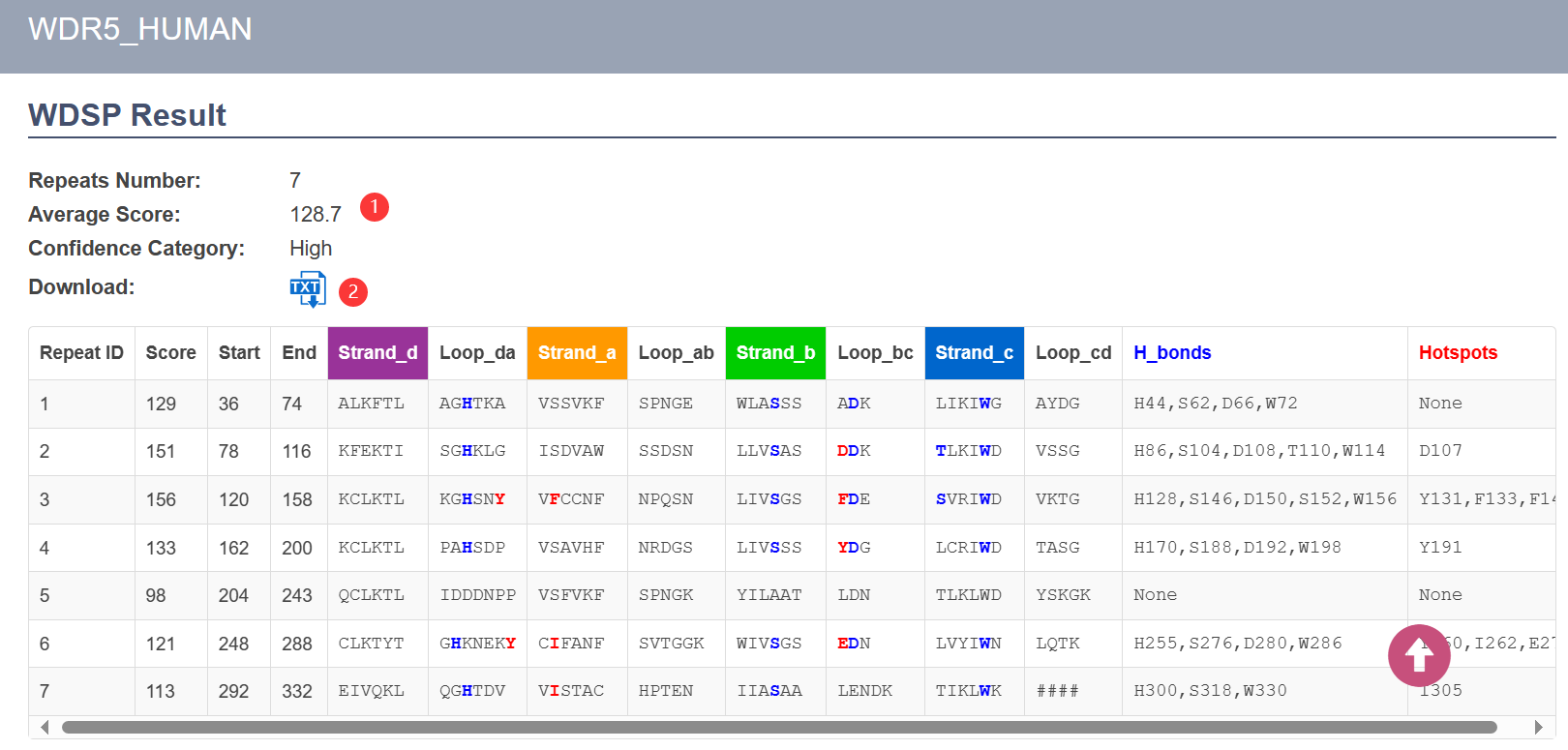
- The WD40 repeats and secondary structures were predicted by the updated WDSP. On top of the table, there are the predicted repeats number, avearge repeat score, and the assigned confidence category ①
- You can download the secondary structures and hydrogen-bond and hotspot residues in text format ②.
- Each line in the colored table below is a single WD40 repeat. The columns are described in the following:
| Column | Explanation |
|---|---|
| Repeat ID | The WD40 repeat order number |
| Score | The score of the predicted WD40 repeat given by WDSP |
| Start | The start site of the WD40 repeat, means the first site of the strand_d |
| End | 'End' here means the first site of the loop_cd of this repeat. For the last repeat, the 'End' site is the last site of strand_c of this repeat, because the last repeat has no loop_cd |
| Strand_d | The first strand of the WD40 repeat at the side face of the structure |
| Loop_da | The loop connecting the strand_d and strand_a at the top-side face of the structure |
| Strand_a | The second strand of the WD40 repeat at the inner face of the structure |
| Loop_ab | The loop connecting the strand_a and strand_b at the bottom face of the structure |
| Strand_b | The third strand of the WD40 repeat |
| Loop_bc | The loop connecting the strand_b and strand_c at the top face of the structure |
| Strand_c | The fourth strand of the WD40 repeat |
| Loop_cd | The loop connecting the strand_c and strand_d of next WD40 repeat at the side-bottom face of the structure |
| H_bonds | The residues participate in forming hydrogen bond networks of WD40 repeat (also colored blue) |
| Hotspots | The potential hotspost residues on the top face (also colored red) |
Structure model & Variants (if mapped)
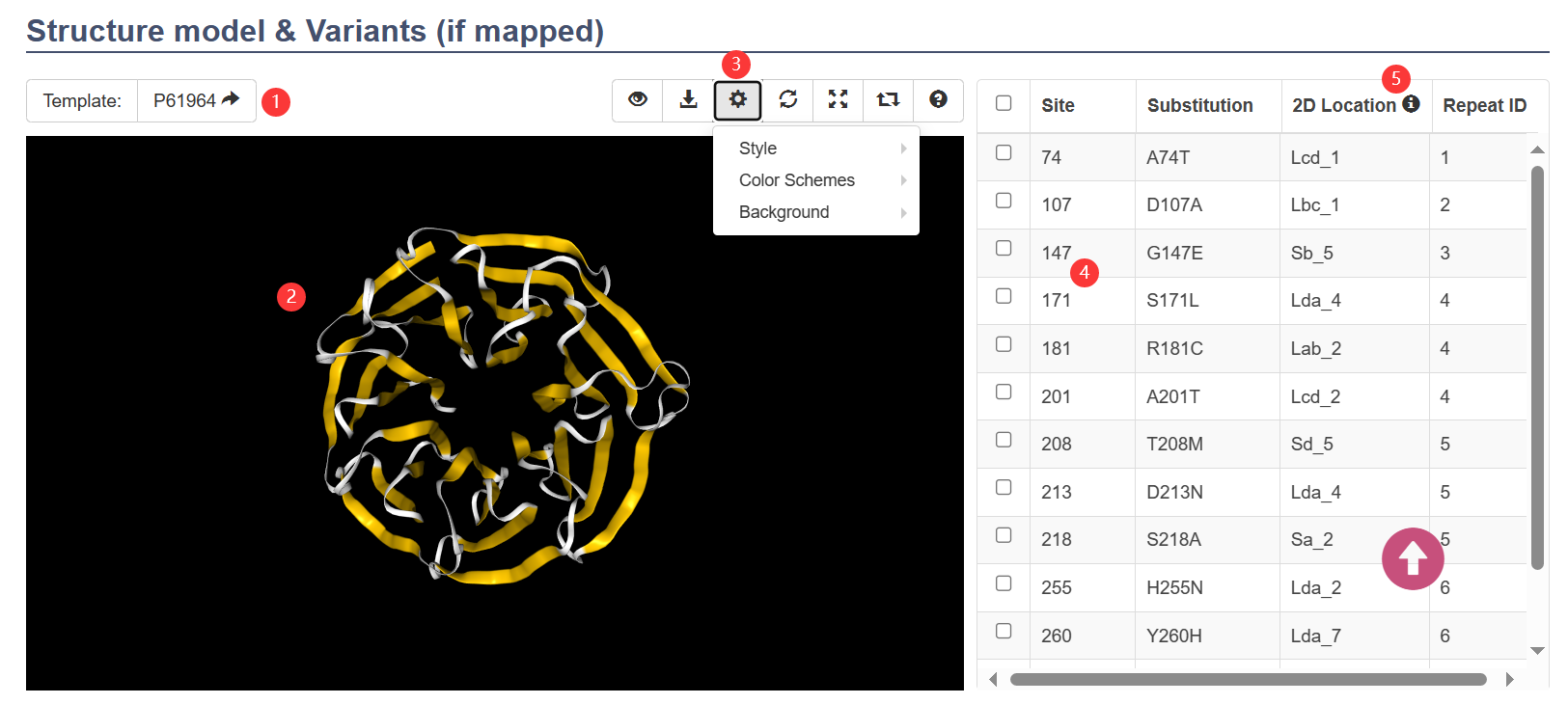
- Only the proteins whose repeat number are 6, 7, or 8, and the whose confidence categories are 'High' have structure models. All of these structure models are modelled using Modeller by adopting a customized alignment method.
- When the template is itself, this means that this model was built by using itself's experimental structure as the template ①.
- When the template is another UniProt Accession Number, this means the structure model was built by using another protein's structure model (also computaionally built) as the template.
- The secondary structures of the 3D structure models have been annotated by using the WDSP's predicted secondary structures. In the default color scheme, the yellow indicated the β-strand, the magenta indicated the α helix and the white indicated the loops ②.
- You can click the buttons on the action bar to view featured sites, download the 3D structure models or associated sequences, and change the structural display style ③.
- Additionally, you can check the specific variants in the checkbox table at the right side to display ④.
- The display color: sites of variants is grey; sites of hydrogen bond networks is blue; sites of variants on hydrogen bond networks is cyan; sites of potential hotspots is red; sites of variants on hotspots is pink;
- ⑤ The secondary structure locations of the variants in the WD40 repeats. For example: "Sa_6" means the sixth residue of strand a; "Lda_7" means the seventh residue of loop da.
| Button | Explanation |
|---|---|
| Template | Jump to the template page |
| View the featured sites as sticks, including potential hotspots on the top face and hydrogen bond networks | |
| Download the 3D structure models or associated sequences | |
| Change the structural display style: style, color schemes or background | |
| Reset all actions | |
| Expand to full screen to view | |
| Let the structure spin | |
| Jump to the help page of 3D structure viewer |
The variants table
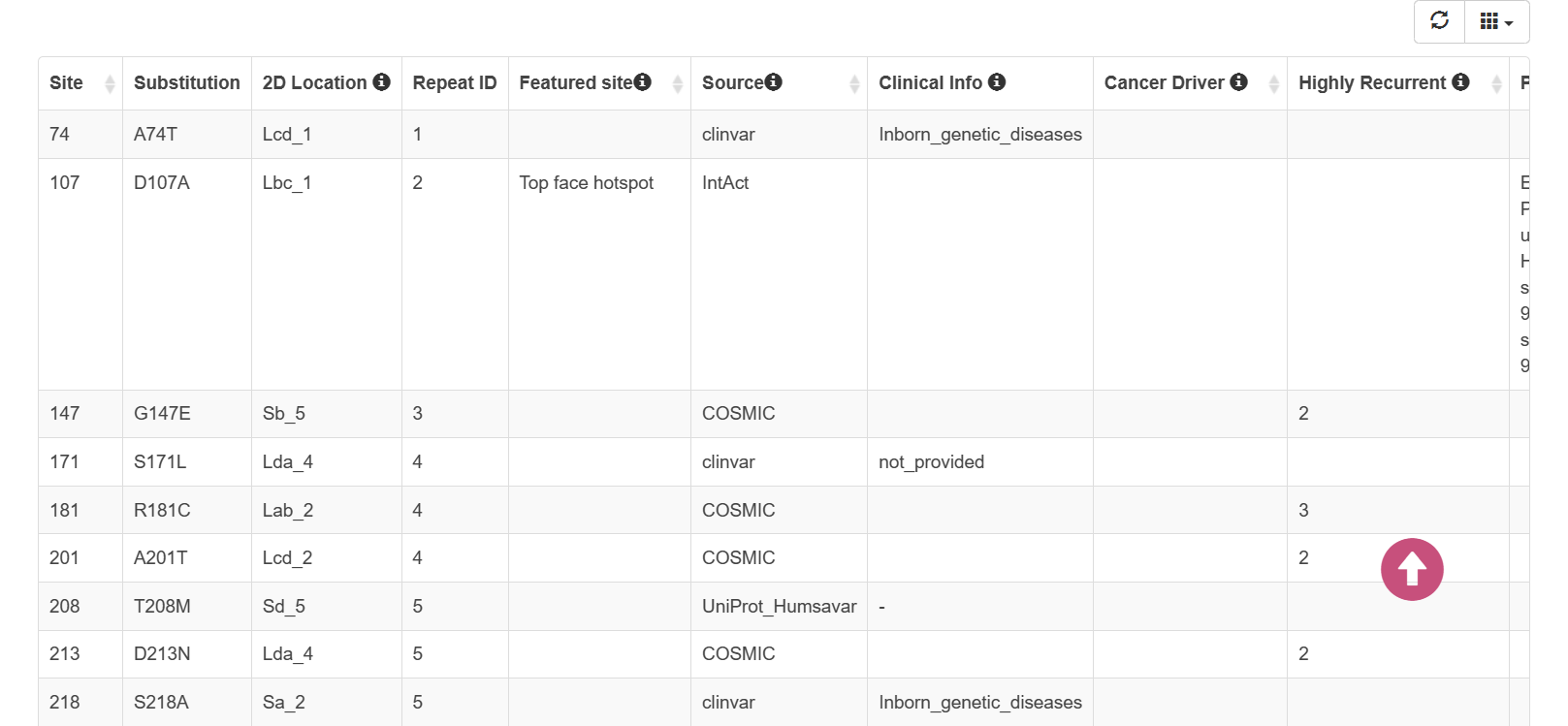
- The table showed the missense variants of human WD40 proteins that belong to “High” confidence category and "Swiss-Prot" section.
- These mutations/variants were collected from different datasets, include IntOGen, IntAct, Clinvar, Cosmic, CancerHotspots, Humsavar, 1000 Genomes Phase 4, ExAC, gnomAD.
- You can select and show specific columns about the mutations/variants and sort them in the dynamic table.
| Display by default | Column | Explanation |
|---|---|---|
| On | Site | Variant site of the UniProt sequence |
| On | Substitution | The single amino acid substitution of the variant |
| On | 2D Location | The location of variant on the secondary structure |
| On | Repeat ID | The ordinal number of WD40 repeat where the variant are located |
| On | Featured site | The variant is on the hydrogen bond network or hotspot residues on the top face |
| On | Resource | The data source or database of the variant |
| On | Clinical Info | The clinical information of the variant from ClinVar, Humsavar, or Cosmic. Please see here. |
| On | Cancer Driver | The associated cancer types driven by this variant, as annotated in IntOGen.Please see here. |
| On | Highly Recurrent | Whether the variant is highly recurrent in cancer, as annotated in CancerHotspots or recurrent in COSMIC. |
| On | PPI Effect | The variant effect on the interaction and specific partner, as annotated in the IntAct. Please see. |
| On | Humsavar Category | For variants from Humsavar of Swiss-Prot, a category (LP/P, LB/B, US) is provided. Please see here . |
| On | ClinVAR Significance | For variants from ClinVAR, the significance is provided, as annotated in ClinVAR. |
| Off | Humsavar FTid | For variants from Humsavar, the Humsavar FTid is provided. |
| Off | ClinVAR ID | For variants from ClinVAR, the ClinVAR ID is provided. |
| Off | ClinVAR Review Status | For variants from ClinVAR, the ClinVAR Review Status is provided. |
| Off | Genome Assembly | If available, the genome assembly that the variant was mapped is provided |
| Off | Chromosome Coordinate | If available, the genome coordinate that the variant was mapped to the specific genome assembly is provided. |
| Off | Reference | If available, the PMID of the reference which reported the variant |
| Off | Allele Frequency | The allele frequency of the variant from the dataset |
| Off | Reference CDS Changes | The transcript codon changes of the variant |
| Off | SIFT Class | Prediction of SIFT: "T(olerated)" or "D(eleterious)", The score cutoff between "D" and "T" is 0.05. |
| Off | SIFT Score | Sorting Intolerant From Tolerant score, range from 0 to 1 |
| Off | MutationAssessor Class | MutationAssessor's functional impact of a variant : predicted functional, i.e. high ("H") or medium ("M"), or predicted non-functional, i.e. low ("L") or neutral ("N"). The MAori score cutoffs between "H" and "M", "M" and "L", and "L" and "N", are 3.5, 1.935 and 0.8, respectively. |
| Off | MutationAssessor Score | MutationAssessor functional impact combined score, The score ranges from -5.135 to 6.49 in dbNSFP. |
| Off | FATHMM Class | If a FATHMM score is <=-1.5 (or rankscore >=0.81332) the corresponding nsSNV is predicted as "D(AMAGING)"; otherwise it is predicted as "T(OLERATED)". |
| Off | FATHMM Score | FATHMM default score (weighted for human inherited-disease mutations with Disease Ontology) (FATHMMori). Scores range from -16.13 to 10.64. The smaller the score the more likely the SNP has damaging effect. |
| Off | MetaSVM class | Prediction of our SVM based ensemble prediction score,"T(olerated)" or "D(amaging)". The score cutoff between "D" and "T" is 0. |
| Off | MetaSVM Score | Support vector machine (SVM) based ensemble prediction score, which incorporated 10 scores (SIFT, PolyPhen-2 HDIV, PolyPhen-2 HVAR, GERP++, MutationTaster, Mutation Assessor, FATHMM, LRT, SiPhy, PhyloP) and the maximum frequency observed in the 1000 genomes populations. Larger value means the SNV is more likely to be damaging. Scores range from -2 to 3 in dbNSFP. |
| Off | MetaLR Class | Prediction of MetaLR based ensemble prediction score,"T(olerated)" or "D(amaging)". The score cutoff between "D" and "T" is 0.5. |
| Off | MetaLR Score | Logistic regression (LR) based ensemble prediction score, which incorporated 10 scores (SIFT, PolyPhen-2 HDIV, PolyPhen-2 HVAR, GERP++, MutationTaster, Mutation Assessor, FATHMM, LRT, SiPhy, PhyloP) and the maximum frequency observed in the 1000 genomes populations. Larger value means the SNV is more likely to be damaging. Scores range from 0 to 1. |
| Off | CADD Phred | CADD score.The larger the score the more likely the SNP has damaging effect.If you would like to apply a cutoff on deleteriousness, e.g. to identify potentially pathogenic variants, we would suggest to put a cutoff somewhere between 10 and 20. Maybe at 15, as this also happens to be the median value for all possible canonical splice site changes and non-synonymous variants. However, there is not a natural choice here -- it is always arbitrary. We therefore recommend integrating C-scores with other evidence and to rank your candidates for follow up rather than hard filtering. |
| Off | Polyphen2 HDIV Class | Polyphen2 prediction based on HumDiv, "D" ("probably damaging", HDIV score in [0.957,1]), "P" ("possibly damaging", HDIV score in [0.453,0.956]) and "B" ("benign", HDIV score in [0,0.452] ). Score cutoff for binary classification is 0.5 for HDIV score |
| Off | Polyphen2 HDIV Score | Polyphen2 score based on HumDiv. The score ranges from 0 to 1 |
| Off | Polyphen2 HVAR Class | Polyphen2 prediction based on HumVar, "D" ("probably damaging", HVAR score in [0.909,1]), "P" ("possibly damaging", HVAR in [0.447,0.908]) and "B" ("benign", HVAR score in [0,0.446]). Score cutoff for binary classification is 0.5 for HVAR score |
| Off | Polyphen2 HVAR Class | Polyphen2 score based on HumVar. The score ranges from 0 to 1. |
Clinical Information
ClinVar:Cancer Driver
| Abbreviation | Full Name |
|---|---|
| ALL | acute lymphoid leukemia |
| AML | acute myeloid leukemia |
| BLCA | bladder carcinoma |
| BRCA | breast carcinoma |
| CLL | chronic lymphocytic leukemia |
| CM | cutaneous melanoma |
| COREAD | colorectal adenocarcinoma |
| DLBC | diffuse large B cell lymphoma |
| ESCA | esophageal carcinoma |
| GBM | glioblastoma multiforme |
| HC | hepatic carcinoma |
| HNSC | head and neck squamous cell carcinoma |
| LGG | lower grade glioma |
| LUAD | lung adenocarcinoma |
| LUSC | lung squamous cell carcinoma |
| MB | medulloblastoma |
| MM | multiple myeloma |
| NB | neuroblastoma |
| NSCLC | non small cell lung carcinoma |
| OV | serous ovarian adenocarcinoma |
| PA | pilocytic astrocytoma |
| PAAD | pancreas adenocarcinoma |
| PRAD | prostate adenocarcinoma |
| RCCC | renal clear cell carcinoma |
| SCLC | small cell lung carcinoma |
| STAD | stomach adenocarcinoma |
| THCA | thyroid carcinoma |
| UCEC | uterine corpus endometrioid carcinoma |
PPI Effect
- Mutation (MI:0118): A change in a sequence or structure in comparison to a reference entity due to a insertion, deletion or substitution event. This root term is used when there is a mutation present in a protein and the wild type version has not been tested or shown to interact in the referenced paper.
- Mutation causing an interaction (MI:2227): A change in a sequence or structure in comparison to a reference entity due to a insertion, deletion or substitution event that enables an interaction when compared with the wild-type, which does not interact.
- Mutation decreasing interaction (MI:0119): Region of a molecule whose mutation or deletion decreases significantly interaction strength or rate (in the case of interactions inferred from enzymatic reaction).
- Mutation decreasing interaction rate (MI:1130): Region of a molecule whose mutation or deletion decreases significantly interaction rate (in the case of interactions inferred from enzymatic reaction).
- Mutation decreasing interaction strength (MI:1133): Region of a molecule whose mutation or deletion decreases significantly interaction strength.
- Mutation disrupting interaction (MI:0573): Region of a molecule whose mutation or deletion totally disrupts an interaction strength or rate (in the case of interactions inferred from enzymatic reaction).
• Mutation disrupting interaction rate (MI:1129): Region of a molecule whose mutation or deletion totally disrupts an interaction rate (in the case of interactions inferred from enzymatic reaction).
• Mutation disrupting interaction strength (MI:1128): Region of a molecule whose mutation or deletion totally disrupts an interaction strength.
- Mutation increasing interaction (MI:0382): Region of a molecule whose mutation or deletion increases significantly interaction strength or rate (in the case of interactions inferred from enzymatic reaction).
- Mutation increasing interaction rate (MI:1131): Region of a molecule whose mutation or deletion increases significantly interaction rate (in the case of interactions inferred from enzymatic reaction).
- Mutation increasing interaction strength (MI:1132): Region of a molecule whose mutation or deletion increases significantly interaction strength.
- Mutation with no effect (MI:2226): A change in a sequence or structure in comparison to a reference entity due to a insertion, deletion or substitution event that does not have any effect over an interaction when compared with the wild-type.
Predictor Help
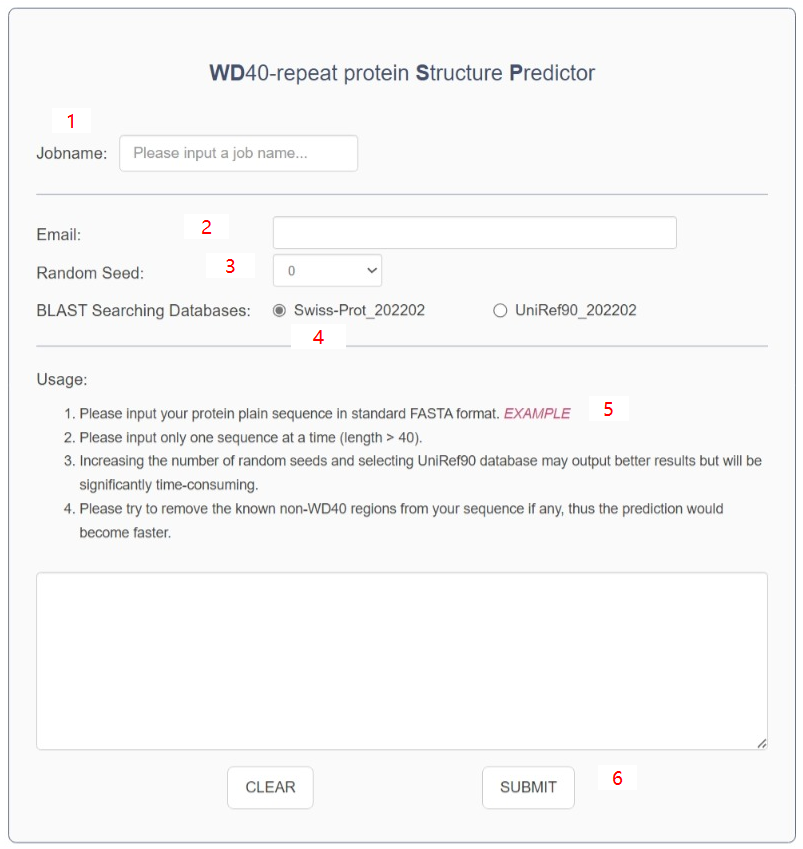
- You should input a jobname to name your submitted task. Empty is not allowed ①.
- You should input your email address ② to receive the link to your prediction result page.
- You can choose different random number generating seeds ③ . The output result only presents the best one in all of your input seeds.
- There are two BLAST searching databases ④: Swiss-Prot 202202 or UniRef90 202202. You could choose the much larger database, UniRef90, to get better predictions. But choosing the UniRef90 would be very time consuming.
- The input sequence must be in standard FASTA format.
- Only one sequence will be processed in each run. And the sequence should be longer than 40. You can click the “EXAMPLE” ⑤ to get the example input.
- Please try to cut the known non-WD40 regions away from your sequence, thus the prediction would become faster.
- Click the “SUBMIT” button ⑥ to submit your task and it would jump to the result page when the task finished. In addition, your email box will receive the result link if provided.
Citations
If you use the data of WDSPdb or the WDSP predictor in your research, please cite the following references:
- Ma, J., An, K., Zhou, J.B., Wu, N.S., Wang, Y., Ye, Z.Q., Wu, Y.D. (2019). WDSPdb: an updated resource for WD40 proteins. Bioinformatics, [PMID: 31161214]
- Wang, Y., Hu, X.J., Zou, X.D., Wu, X.H., Ye, Z.Q. and Wu, Y.D. (2015) WDSPdb: a database for WD40-repeat proteins. Nucleic Acids Res, 43 , D339-344. [PMID: 25348404]
- Wang, Y., Jiang, F., Zhuo, Z., Wu, X.H., Wu, Y.D. (2013). A Method for WD40 Repeat Detection and Secondary Structure Prediction. PLoS ONE , 8(6) , e65705. [PMID: 23776530]
- Wu, X.H., Wang, Y., Zhuo, Z.,Jiang, F., Wu, Y.D. (2012). Identifying the hotspots on the top faces of WD40-repeat proteins from their primary sequences by beta-bulges and DHSW tetrads. PLoS ONE , 7(8) , e43005. [PMID: 22916195]
Frequently Asked Questions
Question List:
A. In the home page, WDSPdb 3.0 supply a search box. It includes two parts: an input box for the keyword used to searching and a dropdown menu for types the keyword belong to. There are 6 types of keywords (UniProt ID, UniProt AC, Gene Name, Entrez ID, Organism, Taxonomy). Besides them, the default of type is “All”, that means it will search your keyword in all 6 types, which may be a little bit slower.
Theoretically, you can use any string as keyword. But we have to remind you to input a keyword as short and accurate as possible in case you cannot get any result. Furthermore, please don’t use “AND” and “:” in your keyword, because they are reserved words of the web site. WDSPdb 3.0 would remove them from your keyword if you input them “carelessly”. Finally, if you input an empty string as keyword, WDSPdb 3.0 will return all entries no matter what type you select. Please don’t do this unless it is necessary.
To help you enter accurate keywords, the input box provides candidate keywords auto-complete function. When you are typing, the box will provides you with possible keywords based on the type you select. For example, if you select the “Organism” type and type “human”, it will provide you with 10 words as candidate keywords, such as “Homo sapiens (human)”. Please choose the candidate keywords as input as possible so that you can get more accurate results. It is important to note that it does not provide you with anything when you select “All” as type.
In addition, we also provide advanced search function. When you click “Add” on the right of “Search” button, below the origin search box a new search box will be added, which also contains the input box and dropdown menu. You can have up to 5 search boxes. Each search box represents a search condition, and they are combined by the logic operator “AND”.
A. The time cost mainly depends on the searching database especially. Generally, if the database is Swiss-Prot, several minutes are sufficient for a protein with about 300 residues; but if the database is Uniref90, it may cost more than half an hour. It also depends on the job queue status. If several jobs are in the queue before your job, you may need to wait for a while. The number of random seeds also affects the running time, the more seeds, the more time needed.
A. After you set search conditions and click the “Search” button, WDSPdb 3.0 will display your search result as a table. There is a button like “” at the top right of the table, which can be clicked to export table content into various formats, such as .json, .xml, .csv, .txt, .sql and so on.
At the top left of the table, there is a dropdown menu, which contains two items: “Export Basic” and “Export Selected”. When you choose “Export Basic”, the export button will export the whole current page of this table. By default, each page of this table contains 25 entries. You can also change it to 10, 50, 100 or all entries of your search result by a dropdown menu at the bottom left of the table. In addition, each row of the table starts with a selection box. When you choose “Export Selected”, you can export several entries you selected.
A. WDSPdb 3.0 offered a REST API for downloading WDSP predicted result of each protein. The URL format of the API is as follows:
www.wdspdb.com/wdspdb3/detail/{UniProt AC}.txt
You can replace {UniProt AC} with a specific Accession Number of a protein in UniProt database. And WDSPdb 3.0 will return the WDSP predicted result of this protein. For example, if you want to get the predicted result of WDR5_HUMAN whose Accession Number is "P61964", then your URL is www.wdspdb.com/wdspdb3/detail/P61964.txt. Don’t forget “.txt” at the end.The protein you focus on must be included in WDSPdb 3.0. Therefore, we recommend that you export a list of proteins from the search results page before using this API. You can call the API programmatically to get results, but please note that we limit the API to only two requests per second to avoid that server load becomes too high.
A. WDSPdb 3.0 is compatible to most modern web browsers, like Chrome, Firefox, Microsoft Edge, and Safari. If your browser does not display the site correctly, please check weather all components have been loaded because sometimes the user's network may encounter some problems. Please do not browse the website in IE browser, because the plug-in we use is not compatible with it. In addition, if you find any bug, please click "Contact" on the top to tell us.